Mostrar el registro sencillo del ítem
dc.contributor.author
Scardino, Valeria
dc.contributor.author
Di Filippo, Juan Ignacio

dc.contributor.author
Cavasotto, Claudio Norberto

dc.date.available
2024-03-04T12:06:25Z
dc.date.issued
2023-01
dc.identifier.citation
Scardino, Valeria; Di Filippo, Juan Ignacio; Cavasotto, Claudio Norberto; How good are AlphaFold models for docking-based virtual screening?; Cell Press; iScience; 26; 1; 1-2023; 1-18
dc.identifier.issn
2589-0042
dc.identifier.uri
http://hdl.handle.net/11336/229198
dc.description.abstract
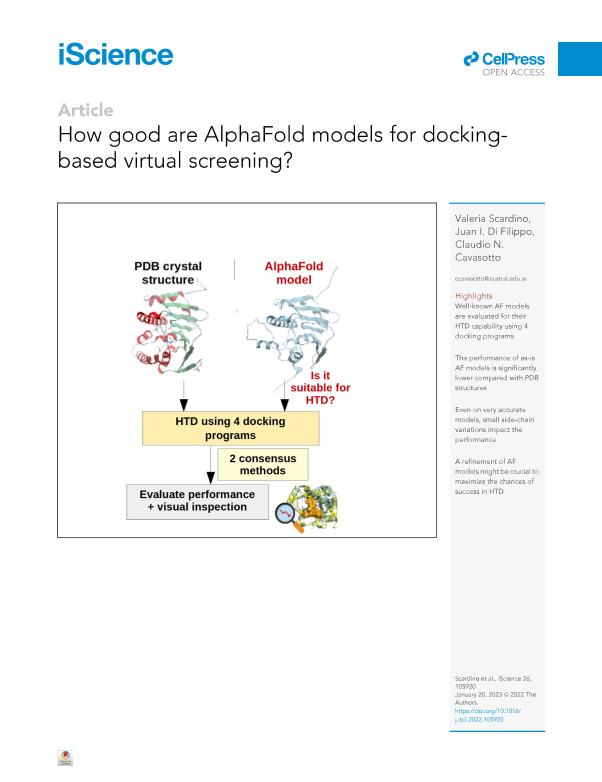
A crucial component in structure-based drug discovery is the availability of high-quality three-dimensional structures of the protein target. Whenever experimental structures were not available, homology modeling has been, so far, the method of choice. Recently, AlphaFold (AF), an artificial-intelligence-based protein structure prediction method, has shown impressive results in terms of model accuracy. This outstanding success prompted us to evaluate how accurate AF models are from the perspective of docking-based drug discovery. We compared the high-throughput docking (HTD) performance of AF models with their corresponding experimental PDB structures using a benchmark set of 22 targets. The AF models showed consistently worse performance using four docking programs and two consensus techniques. Although AlphaFold shows a remarkable ability to predict protein architecture, this might not be enough to guarantee that AF models can be reliably used for HTD, and post-modeling refinement strategies might be key to increase the chances of success.
dc.format
application/pdf
dc.language.iso
eng
dc.publisher
Cell Press

dc.rights
info:eu-repo/semantics/openAccess
dc.rights.uri
https://creativecommons.org/licenses/by-nc-nd/2.5/ar/
dc.subject
ARTIFICIAL INTELLIGENCE
dc.subject
COMPUTATIONAL CHEMISTRY
dc.subject
PROTEIN
dc.subject
PROTEIN FOLDING
dc.subject.classification
Otras Ciencias Químicas

dc.subject.classification
Ciencias Químicas

dc.subject.classification
CIENCIAS NATURALES Y EXACTAS

dc.title
How good are AlphaFold models for docking-based virtual screening?
dc.type
info:eu-repo/semantics/article
dc.type
info:ar-repo/semantics/artículo
dc.type
info:eu-repo/semantics/publishedVersion
dc.date.updated
2024-02-29T12:59:36Z
dc.journal.volume
26
dc.journal.number
1
dc.journal.pagination
1-18
dc.journal.pais
Estados Unidos

dc.description.fil
Fil: Scardino, Valeria. Universidad Austral; Argentina
dc.description.fil
Fil: Di Filippo, Juan Ignacio. Universidad Austral. Facultad de Ciencias Biomédicas. Instituto de Investigaciones en Medicina Traslacional. Consejo Nacional de Investigaciones Científicas y Técnicas. Oficina de Coordinación Administrativa Parque Centenario. Instituto de Investigaciones en Medicina Traslacional; Argentina
dc.description.fil
Fil: Cavasotto, Claudio Norberto. Universidad Austral. Facultad de Ciencias Biomédicas. Instituto de Investigaciones en Medicina Traslacional. Consejo Nacional de Investigaciones Científicas y Técnicas. Oficina de Coordinación Administrativa Parque Centenario. Instituto de Investigaciones en Medicina Traslacional; Argentina. Universidad Austral; Argentina
dc.journal.title
iScience
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/url/https://www.sciencedirect.com/science/article/pii/S2589004222021939
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/doi/http://dx.doi.org/10.1016/j.isci.2022.105920
Archivos asociados