Mostrar el registro sencillo del ítem
dc.contributor.author
Huang, Tsun Tsao
dc.contributor.author
Marcos, María Laura

dc.contributor.author
Hwang, Jenn Kang
dc.contributor.author
Echave, Julián

dc.date.available
2018-01-18T15:04:04Z
dc.date.issued
2014-04
dc.identifier.citation
Huang, Tsun Tsao; Marcos, María Laura; Hwang, Jenn Kang; Echave, Julián; A mechanistic stress model of protein evolution accounts for site-specific evolutionary rates and their relationship with packing density and flexibility; BioMed Central; BMC Evolutionary Biology; 14; 78; 4-2014; 1-9
dc.identifier.issn
1471-2148
dc.identifier.uri
http://hdl.handle.net/11336/33774
dc.description.abstract
BACKGROUND: Protein sites evolve at different rates due to functional and biophysical constraints. It is usually considered that the main structural determinant of a site’s rate of evolution is its Relative Solvent Accessibility (RSA). However, a recent comparative study has shown that the main structural determinant is the site’s Local Packing Density (LPD). LPD is related with dynamical flexibility, which has also been shown to correlate with sequence variability. Our purpose is to investigate the mechanism that connects a site’s LPD with its rate of evolution.
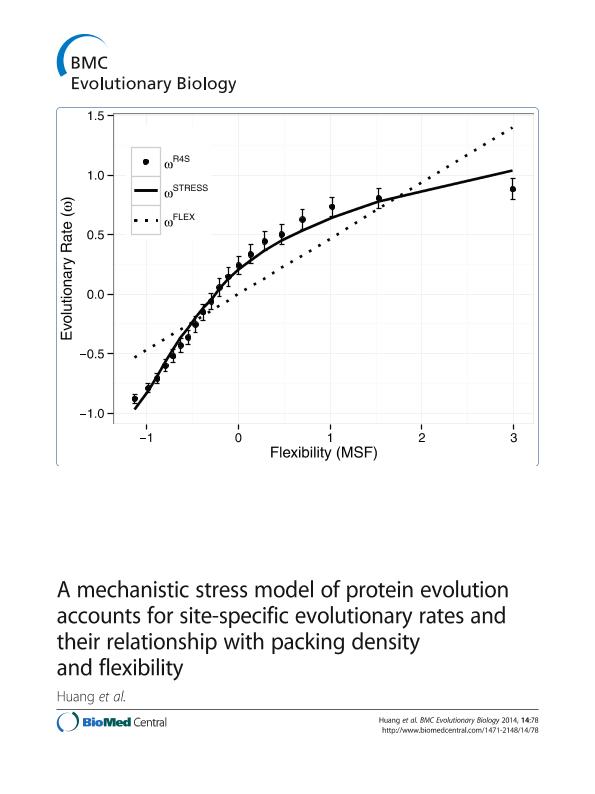
RESULTS: We consider two models: an empirical Flexibility Model and a mechanistic Stress Model. The Flexibility Model postulates a linear increase of site-specific rate of evolution with dynamical flexibility. The Stress Model, introduced here, models mutations as random perturbations of the protein’s potential energy landscape, for which we use simple Elastic Network Models (ENMs). To account for natural selection we assume a single active conformation and use basic statistical physics to derive a linear relationship between site-specific evolutionary rates and the local stress of the mutant’s active conformation. We compare both models on a large and diverse dataset of enzymes. In a protein-by-protein study we found that the Stress Model outperforms the Flexibility Model for most proteins. Pooling all proteins together we show that the Stress Model is strongly supported by the total weight of evidence. Moreover, it accounts for the observed nonlinear dependence of sequence variability on flexibility. Finally, when mutational stress is controlled for, there is very little remaining correlation between sequence variability and dynamical flexibility.
CONCLUSIONS: We developed a mechanistic Stress Model of evolution according to which the rate of evolution of a site is predicted to depend linearly on the local mutational stress of the active conformation. Such local stress is proportional to LPD, so that this model explains the relationship between LPD and evolutionary rate. Moreover, the model also accounts for the nonlinear dependence between evolutionary rate and dynamical flexibility.
dc.format
application/pdf
dc.language.iso
eng
dc.publisher
BioMed Central

dc.rights
info:eu-repo/semantics/openAccess
dc.rights.uri
https://creativecommons.org/licenses/by/2.5/ar/
dc.subject
Protein Evolution
dc.subject
Site-Specific Substitution Rate
dc.subject
Local Packing Density
dc.subject
Elastic Network Model
dc.subject
Flexibility
dc.subject
Stress
dc.subject
Mean Square Fluctuation
dc.subject
Mean Local Mutational Stress
dc.subject.classification
Otras Ciencias Químicas

dc.subject.classification
Ciencias Químicas

dc.subject.classification
CIENCIAS NATURALES Y EXACTAS

dc.title
A mechanistic stress model of protein evolution accounts for site-specific evolutionary rates and their relationship with packing density and flexibility
dc.type
info:eu-repo/semantics/article
dc.type
info:ar-repo/semantics/artículo
dc.type
info:eu-repo/semantics/publishedVersion
dc.date.updated
2018-01-16T18:03:06Z
dc.journal.volume
14
dc.journal.number
78
dc.journal.pagination
1-9
dc.journal.pais
Reino Unido

dc.journal.ciudad
Londres
dc.description.fil
Fil: Huang, Tsun Tsao. National Chiao Tung University; República de China
dc.description.fil
Fil: Marcos, María Laura. Universidad Nacional de San Martín. Escuela de Ciencia y Tecnología; Argentina. Consejo Nacional de Investigaciones Científicas y Técnicas; Argentina
dc.description.fil
Fil: Hwang, Jenn Kang. National Chiao Tung University; República de China
dc.description.fil
Fil: Echave, Julián. Universidad Nacional de San Martín. Escuela de Ciencia y Tecnología; Argentina. Consejo Nacional de Investigaciones Científicas y Técnicas; Argentina
dc.journal.title
BMC Evolutionary Biology

dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/url/https://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-14-78
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/doi/http://dx.doi.org/10.1186/1471-2148-14-78
Archivos asociados