Artículo
Analysis workflow of publicly available RNA-sequencing datasets
Sanchis, Pablo Antonio ; Lavignolle, Rosario; Abbate, María Mercedes
; Lavignolle, Rosario; Abbate, María Mercedes ; Lage Vickers, Sofia
; Lage Vickers, Sofia ; Vazquez, Elba Susana
; Vazquez, Elba Susana ; Cotignola, Javier Hernan
; Cotignola, Javier Hernan ; Bizzotto, Juan Antonio
; Bizzotto, Juan Antonio ; Gueron, Geraldine
; Gueron, Geraldine
 ; Lavignolle, Rosario; Abbate, María Mercedes
; Lavignolle, Rosario; Abbate, María Mercedes ; Lage Vickers, Sofia
; Lage Vickers, Sofia ; Vazquez, Elba Susana
; Vazquez, Elba Susana ; Cotignola, Javier Hernan
; Cotignola, Javier Hernan ; Bizzotto, Juan Antonio
; Bizzotto, Juan Antonio ; Gueron, Geraldine
; Gueron, Geraldine
Fecha de publicación:
06/2021
Editorial:
Cell Press
Revista:
STAR Protocols
ISSN:
2666-1667
Idioma:
Inglés
Tipo de recurso:
Artículo publicado
Clasificación temática:
Resumen
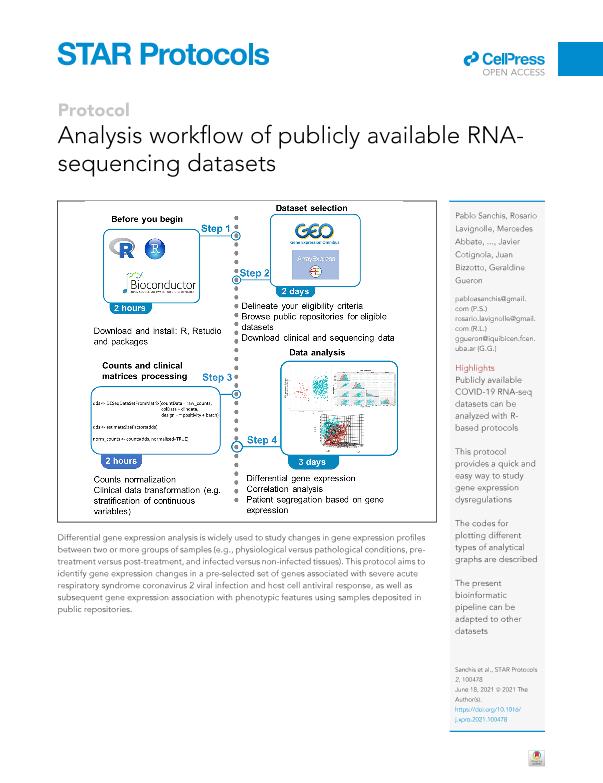
Differential gene expression analysis is widely used to study changes in gene expression profiles between two or more groups of samples (e.g., physiological versus pathological conditions, pre-treatment versus post-treatment, and infected versus non-infected tissues). This protocol aims to identify gene expression changes in a pre-selected set of genes associated with severe acute respiratory syndrome coronavirus 2 viral infection and host cell antiviral response, as well as subsequent gene expression association with phenotypic features using samples deposited in public repositories. For complete details on the use and outcome of this informatics analysis, please refer to Bizzotto et al. (2020).
Palabras clave:
BIOINFORMATICS
,
RNASEQ
Archivos asociados
Licencia
Identificadores
Colecciones
Articulos(IQUIBICEN)
Articulos de INSTITUTO DE QUIMICA BIOLOGICA DE LA FACULTAD DE CS. EXACTAS Y NATURALES
Articulos de INSTITUTO DE QUIMICA BIOLOGICA DE LA FACULTAD DE CS. EXACTAS Y NATURALES
Citación
Sanchis, Pablo Antonio; Lavignolle, Rosario; Abbate, María Mercedes; Lage Vickers, Sofia; Vazquez, Elba Susana; et al.; Analysis workflow of publicly available RNA-sequencing datasets; Cell Press; STAR Protocols; 2; 2; 6-2021; 1-19
Compartir
Altmétricas