Mostrar el registro sencillo del ítem
dc.contributor.author
Brahimi, Fouad
dc.contributor.author
Galan, Alba
dc.contributor.author
Jmaeff, Sean
dc.contributor.author
Barcelona, Pablo Federico

dc.contributor.author
De Jay, Nicolas
dc.contributor.author
Dejgaard, Kurt
dc.contributor.author
Young, Jason C.
dc.contributor.author
Kleinman, Claudia Laura

dc.contributor.author
Thomas, David Y.
dc.contributor.author
Saragovi, H. Uri
dc.date.available
2022-06-01T14:03:50Z
dc.date.issued
2020-08
dc.identifier.citation
Brahimi, Fouad; Galan, Alba; Jmaeff, Sean; Barcelona, Pablo Federico; De Jay, Nicolas; et al.; Alternative splicing of a receptor Intracellular domain yields different ectodomain conformations, enabling Isoform-Selective Functional Ligands; Bellwether Publ Ltd; Giscience & Remote Sensing; 23; 9; 8-2020; 1-34
dc.identifier.issn
1548-1603
dc.identifier.uri
http://hdl.handle.net/11336/158658
dc.description.abstract
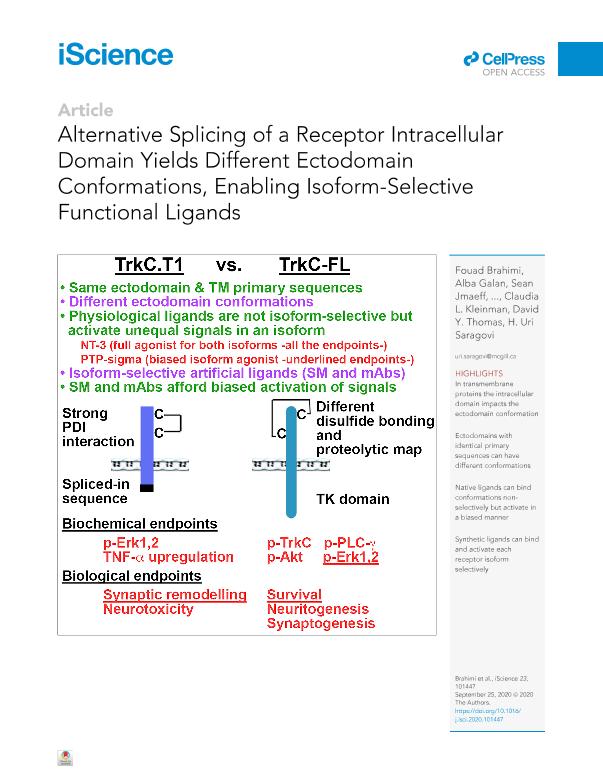
Events at a receptor ectodomain affect the intracellular domain conformation, activating signal transduction (out-to-in conformational effects). We investigated there verse direction (in-to-out) where the intracellular domain may impact on ectodomain conformation. The primary sequences of naturally occurring TrkC receptor isoforms(TrkC-FL and TrkC.T1) only differ at the intracellular domain. However, owing to their differential association with Protein Disulfide Isomerase the isoforms have different disulfide bonding and conformations at the ectodomain. Conformations were exploited to develop artificial ligands, mAbs, and small molecules, with isoform-specific binding and biased activation. Consistent, the physiological ligandsNT-3 and PTP-sigma bind both isoforms, but NT-3 activates all signaling pathways,whereas PTP-sigma activates biased signals. Our data support an "in-to-out" model controlling receptor ectodomain conformation, a strategy that enables heterogeneityin receptors, ligands, and bioactivity. These concepts may be extended to the many wild-type or oncogenic receptors with known isoforms.
dc.format
application/pdf
dc.language.iso
eng
dc.publisher
Bellwether Publ Ltd

dc.rights
info:eu-repo/semantics/openAccess
dc.rights.uri
https://creativecommons.org/licenses/by-nc-sa/2.5/ar/
dc.subject
TrkC-FL
dc.subject
TrkC.T1
dc.subject.classification
Bioquímica y Biología Molecular

dc.subject.classification
Ciencias Biológicas

dc.subject.classification
CIENCIAS NATURALES Y EXACTAS

dc.title
Alternative splicing of a receptor Intracellular domain yields different ectodomain conformations, enabling Isoform-Selective Functional Ligands
dc.type
info:eu-repo/semantics/article
dc.type
info:ar-repo/semantics/artículo
dc.type
info:eu-repo/semantics/publishedVersion
dc.date.updated
2021-09-06T15:11:09Z
dc.identifier.eissn
1943-7226
dc.journal.volume
23
dc.journal.number
9
dc.journal.pagination
1-34
dc.journal.pais
Estados Unidos

dc.description.fil
Fil: Brahimi, Fouad. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Galan, Alba. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Jmaeff, Sean. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Barcelona, Pablo Federico. Mc Gill University. Lady Davis Research Intitute; Canadá. Consejo Nacional de Investigaciones Científicas y Técnicas. Centro Científico Tecnológico Córdoba. Centro de Investigaciones en Bioquímica Clínica e Inmunología; Argentina
dc.description.fil
Fil: De Jay, Nicolas. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Dejgaard, Kurt. McGill University; Canadá
dc.description.fil
Fil: Young, Jason C.. McGill University; Canadá
dc.description.fil
Fil: Kleinman, Claudia Laura. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Thomas, David Y.. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.description.fil
Fil: Saragovi, H. Uri. Mc Gill University. Lady Davis Research Intitute; Canadá
dc.journal.title
Giscience & Remote Sensing

dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/doi/https://doi.org/10.1016/j.isci.2020.101447
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/url/https://www.cell.com/iscience/fulltext/S2589-0042(20)30639-8
Archivos asociados