Artículo
Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer
Kugel, Sita; Feldman, Jessica L.; Klein, Mark A.; Silberman, Dafne Magali ; Sebastián, Carlos; Mermel, Craig; Dobersch, Stephanie; Clark, Abbe R.; Getz, Gad; Denu, John M.; Mostoslavsky, Raul
; Sebastián, Carlos; Mermel, Craig; Dobersch, Stephanie; Clark, Abbe R.; Getz, Gad; Denu, John M.; Mostoslavsky, Raul
 ; Sebastián, Carlos; Mermel, Craig; Dobersch, Stephanie; Clark, Abbe R.; Getz, Gad; Denu, John M.; Mostoslavsky, Raul
; Sebastián, Carlos; Mermel, Craig; Dobersch, Stephanie; Clark, Abbe R.; Getz, Gad; Denu, John M.; Mostoslavsky, Raul
Fecha de publicación:
10/2015
Editorial:
Elsevier Inc
Revista:
Cell Reports
ISSN:
2211-1247
Idioma:
Inglés
Tipo de recurso:
Artículo publicado
Clasificación temática:
Resumen
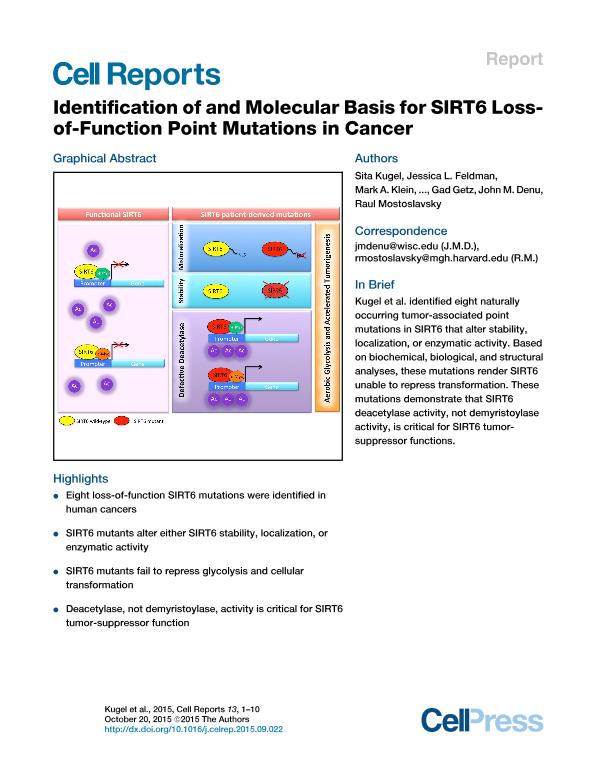
Chromatin factors have emerged as the most frequently dysregulated family of proteins in cancer. We have previously identified the histone deacetylase SIRT6 as a key tumor suppressor, yet whether point mutations are selected for in cancer remains unclear. In this manuscript, we characterized naturally occurring patient-derived SIRT6 mutations. Strikingly, all the mutations significantly affected either stability or catalytic activity of SIRT6, indicating that these mutations were selected for in these tumors. Further, the mutant proteins failed to rescue sirt6 knockout (SIRT6 KO) cells, as measured by the levels of histone acetylation at glycolytic genes and their inability to rescue the tumorigenic potential of these cells. Notably, the main activity affected in the mutants was histone deacetylation rather than demyristoylation, pointing to the former as the main tumor-suppressive function for SIRT6. Our results identified cancer-associated point mutations in SIRT6, cementing its function as a tumor suppressor in human cancer.
Archivos asociados
Licencia
Identificadores
Colecciones
Articulos(CEFYBO)
Articulos de CENTRO DE ESTUDIOS FARMACOLOGICOS Y BOTANICOS
Articulos de CENTRO DE ESTUDIOS FARMACOLOGICOS Y BOTANICOS
Citación
Kugel, Sita ; Feldman, Jessica L. ; Klein, Mark A.; Silberman, Dafne Magali; Sebastián, Carlos; et al.; Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer; Elsevier Inc; Cell Reports; 13; 3; 10-2015; 479-488
Compartir
Altmétricas