Mostrar el registro sencillo del ítem
dc.contributor.author
Monteiro, Lummy Maria Oliveira
dc.contributor.author
Arruda, Leticia Magalhães
dc.contributor.author
Sanches Medeiros, Ananda
dc.contributor.author
Martins Santana, Leonardo
dc.contributor.author
Alves, Luana de Fátima
dc.contributor.author
Defelipe, Lucas Alfredo

dc.contributor.author
Turjanski, Adrian Gustavo
dc.contributor.author
Guazzaroni, Mara Eugenia
dc.contributor.author
de Lorenzo, Victor
dc.contributor.author
Silva Rocha, Rafael
dc.date.available
2021-01-21T12:57:59Z
dc.date.issued
2019-07
dc.identifier.citation
Monteiro, Lummy Maria Oliveira; Arruda, Leticia Magalhães; Sanches Medeiros, Ananda; Martins Santana, Leonardo; Alves, Luana de Fátima; et al.; Reverse Engineering of an Aspirin-Responsive Transcriptional Regulator in Escherichia coli; American Chemical Society; ACS Synthetic Biology; 8; 8; 7-2019; 1890-1900
dc.identifier.issn
2161-5063
dc.identifier.uri
http://hdl.handle.net/11336/123278
dc.description.abstract
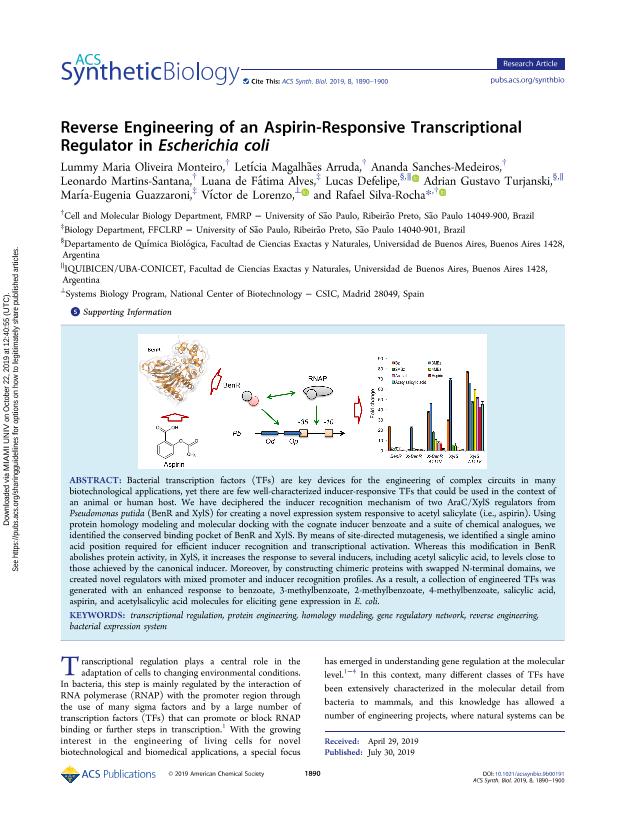
Bacterial transcription factors (TFs) are key devices for the engineering of complex circuits in many biotechnological applications, yet there are few well-characterized inducer-responsive TFs that could be used in the context of an animal or human host. We have deciphered the inducer recognition mechanism of two AraC/XylS regulators from Pseudomonas putida (BenR and XylS) for creating a novel expression system responsive to acetyl salicylate (i.e., aspirin). Using protein homology modeling and molecular docking with the cognate inducer benzoate and a suite of chemical analogues, we identified the conserved binding pocket of BenR and XylS. By means of site-directed mutagenesis, we identified a single amino acid position required for efficient inducer recognition and transcriptional activation. Whereas this modification in BenR abolishes protein activity, in XylS, it increases the response to several inducers, including acetyl salicylic acid, to levels close to those achieved by the canonical inducer. Moreover, by constructing chimeric proteins with swapped N-terminal domains, we created novel regulators with mixed promoter and inducer recognition profiles. As a result, a collection of engineered TFs was generated with an enhanced response to benzoate, 3-methylbenzoate, 2-methylbenzoate, 4-methylbenzoate, salicylic acid, aspirin, and acetylsalicylic acid molecules for eliciting gene expression in E. coli.
dc.format
application/pdf
dc.language.iso
eng
dc.publisher
American Chemical Society

dc.rights
info:eu-repo/semantics/openAccess
dc.rights.uri
https://creativecommons.org/licenses/by-nc-sa/2.5/ar/
dc.subject
bacterial expression system
dc.subject
transcriptional regulation
dc.subject
protein engineering
dc.subject
homology modeling
dc.subject.classification
Bioquímica y Biología Molecular

dc.subject.classification
Ciencias Biológicas

dc.subject.classification
CIENCIAS NATURALES Y EXACTAS

dc.title
Reverse Engineering of an Aspirin-Responsive Transcriptional Regulator in Escherichia coli
dc.type
info:eu-repo/semantics/article
dc.type
info:ar-repo/semantics/artículo
dc.type
info:eu-repo/semantics/publishedVersion
dc.date.updated
2020-12-01T16:25:38Z
dc.identifier.eissn
2161-5063
dc.journal.volume
8
dc.journal.number
8
dc.journal.pagination
1890-1900
dc.journal.pais
Estados Unidos

dc.description.fil
Fil: Monteiro, Lummy Maria Oliveira. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: Arruda, Leticia Magalhães. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: Sanches Medeiros, Ananda. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: Martins Santana, Leonardo. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: Alves, Luana de Fátima. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: Defelipe, Lucas Alfredo. Consejo Nacional de Investigaciones Científicas y Técnicas. Oficina de Coordinación Administrativa Ciudad Universitaria. Instituto de Química Biológica de la Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Instituto de Química Biológica de la Facultad de Ciencias Exactas y Naturales; Argentina. Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Departamento de Química Biológica; Argentina
dc.description.fil
Fil: Turjanski, Adrian Gustavo. Consejo Nacional de Investigaciones Científicas y Técnicas. Oficina de Coordinación Administrativa Ciudad Universitaria. Instituto de Química Biológica de la Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Instituto de Química Biológica de la Facultad de Ciencias Exactas y Naturales; Argentina. Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Departamento de Química Biológica; Argentina
dc.description.fil
Fil: Guazzaroni, Mara Eugenia. Universidade de Sao Paulo; Brasil
dc.description.fil
Fil: de Lorenzo, Victor. Consejo Superior de Investigaciones Científicas. Centro Nacional de Biotecnología; España
dc.description.fil
Fil: Silva Rocha, Rafael. Universidade de Sao Paulo; Brasil
dc.journal.title
ACS Synthetic Biology
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/url/https://pubs.acs.org/doi/10.1021/acssynbio.9b00191
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/doi/http://dx.doi.org/10.1021/acssynbio.9b00191
Archivos asociados