Mostrar el registro sencillo del ítem
dc.contributor.author
Masone, Diego Fernando

dc.contributor.author
Uhart, Marina

dc.contributor.author
Bustos, Diego Martin

dc.date.available
2019-11-29T21:45:09Z
dc.date.issued
2018-04
dc.identifier.citation
Masone, Diego Fernando; Uhart, Marina; Bustos, Diego Martin; Bending Lipid Bilayers: A Closed-Form Collective Variable for Effective Free-Energy Landscapes in Quantitative Biology; American Chemical Society; Journal of Chemical Theory and Computation; 14; 4; 4-2018; 2240-2245
dc.identifier.issn
1549-9618
dc.identifier.uri
http://hdl.handle.net/11336/91051
dc.description.abstract
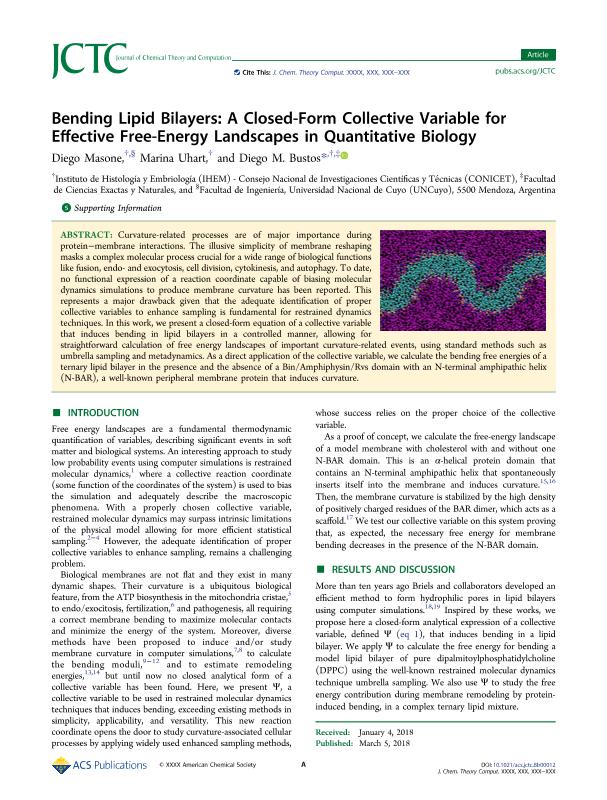
Curvature-related processes are of major importance during protein-membrane interactions. The illusive simplicity of membrane reshaping masks a complex molecular process crucial for a wide range of biological functions like fusion, endo- and exocytosis, cell division, cytokinesis, and autophagy. To date, no functional expression of a reaction coordinate capable of biasing molecular dynamics simulations to produce membrane curvature has been reported. This represents a major drawback given that the adequate identification of proper collective variables to enhance sampling is fundamental for restrained dynamics techniques. In this work, we present a closed-form equation of a collective variable that induces bending in lipid bilayers in a controlled manner, allowing for straightforward calculation of free energy landscapes of important curvature-related events, using standard methods such as umbrella sampling and metadynamics. As a direct application of the collective variable, we calculate the bending free energies of a ternary lipid bilayer in the presence and the absence of a Bin/Amphiphysin/Rvs domain with an N-terminal amphipathic helix (N-BAR), a well-known peripheral membrane protein that induces curvature.
dc.format
application/pdf
dc.language.iso
eng
dc.publisher
American Chemical Society

dc.rights
info:eu-repo/semantics/openAccess
dc.rights.uri
https://creativecommons.org/licenses/by-nc-sa/2.5/ar/
dc.subject
lipid bilayers
dc.subject
Collective Variable
dc.subject
bend
dc.subject
Free-Energy
dc.subject
Free-Energy
dc.subject.classification
Otras Ciencias Naturales y Exactas

dc.subject.classification
Otras Ciencias Naturales y Exactas

dc.subject.classification
CIENCIAS NATURALES Y EXACTAS

dc.title
Bending Lipid Bilayers: A Closed-Form Collective Variable for Effective Free-Energy Landscapes in Quantitative Biology
dc.type
info:eu-repo/semantics/article
dc.type
info:ar-repo/semantics/artículo
dc.type
info:eu-repo/semantics/publishedVersion
dc.date.updated
2019-10-21T20:07:05Z
dc.journal.volume
14
dc.journal.number
4
dc.journal.pagination
2240-2245
dc.journal.pais
Estados Unidos

dc.description.fil
Fil: Masone, Diego Fernando. Consejo Nacional de Investigaciones Científicas y Técnicas. Centro Científico Tecnológico Conicet - Mendoza. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos. Universidad Nacional de Cuyo. Facultad de Ciencias Médicas. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos; Argentina
dc.description.fil
Fil: Uhart, Marina. Consejo Nacional de Investigaciones Científicas y Técnicas. Centro Científico Tecnológico Conicet - Mendoza. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos. Universidad Nacional de Cuyo. Facultad de Ciencias Médicas. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos; Argentina
dc.description.fil
Fil: Bustos, Diego Martin. Consejo Nacional de Investigaciones Científicas y Técnicas. Centro Científico Tecnológico Conicet - Mendoza. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos. Universidad Nacional de Cuyo. Facultad de Ciencias Médicas. Instituto de Histología y Embriología de Mendoza Dr. Mario H. Burgos; Argentina
dc.journal.title
Journal of Chemical Theory and Computation

dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/url/http://pubs.acs.org/doi/10.1021/acs.jctc.8b00012
dc.relation.alternativeid
info:eu-repo/semantics/altIdentifier/doi/http://dx.doi.org/10.1021/acs.jctc.8b00012
Archivos asociados